当前位置:网站首页>医学图像数据增强-归一化
医学图像数据增强-归一化
2022-08-08 13:44:00 【智能之心】
normalizations.py
# Copyright 2021 Division of Medical Image Computing, By DJ.
# German Cancer Research Center (DKFZ)
# https://github.com/MIC-DKFZ/batchgenerators/blob/master/batchgenerators/augmentations/normalizations.py
# and Applied Computer Vision Lab, Helmholtz Imaging Platform
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
r"""
range_normalization(data, per_channel=True, rnge=(0, 1), eps=1e-8)
min_max_normalization(data, eps)
zero_mean_unit_variance_normalization(data, per_channel=True, epsilon=1e-8)
mean_std_normalization(data, mean, std, per_channel=True)
cut_off_outliers(data, percentile_lower=0.2, percentile_upper=99.8, per_channel=False)
-focus:
from normalizations import (range_normalization, min_max_normalization, zero_mean_unit_variance_normalization, mean_std_normalization, cut_off_outliers)
"""
def range_normalization(data, per_channel=True, rnge=(0, 1), eps=1e-8):
r""" 最大最小值归一,默认归一到 (0,1)之间
@Args:
data: (b,c,z(,y(,x)))
rnge: default (0,1)
per_channel: normal in 对每个通道单独进行, 或所有通道同时进行.
@Returns:
data_normalized : shape like data'inputs, dtype=np.float32.
"""
data_normalized = np.zeros(data.shape, dtype=np.float32)
for b in range(data.shape[0]):
if per_channel:
for c in range(data.shape[1]):
tmp_normalized = min_max_normalization(data[b, c], eps)
assert data_normalized.dtype==tmp_normalized.dtype, "Type must be the same!"
data_normalized[b, c] = tmp_normalized
pass
pass
else:
tmp_normalized = min_max_normalization(data[b], eps)
assert data_normalized.dtype==tmp_normalized.dtype, "Type must be the same!"
data_normalized[b] = tmp_normalized
pass
pass
data_normalized *= (rnge[1] - rnge[0])
data_normalized += rnge[0]
return data_normalized
def min_max_normalization(data, eps):
r""" function: (x-a)/(b-a+eps)
@Args:
data: (z(,y(,x)))
"""
mn = data.min()
mx = data.max()
data_normalized = data - mn
old_range = mx - mn + eps
data_normalized = np.true_divide(data_normalized, old_range)
return np.array(data_normalized, dtype=np.float32)
def zero_mean_unit_variance_normalization(data, per_channel=True, epsilon=1e-8):
r"""零均值方差单元归一化
@Args:
data: (b,c,z(,y(,x)))
per_channel: normal in 对每个通道单独进行, 或所有通道同时进行.
@Returns:
data_normalized : shape like data'inputs, dtype=np.float32.
"""
data_normalized = np.zeros(data.shape, dtype=np.float32)
for b in range(data.shape[0]):
if per_channel:
for c in range(data.shape[1]):
mean = data[b, c].mean()
std = data[b, c].std() + epsilon
data_normalized[b, c] = (data[b, c] - mean) / std
pass
pass
else:
mean = data[b].mean()
std = data[b].std() + epsilon
data_normalized[b] = (data[b] - mean) / std
pass
pass
return data_normalized
def mean_std_normalization(data, mean, std, per_channel=True):
r"""自定义方差和标准差归一方法
@Args:
data: (b,c,z(,y(,x)))
per_channel: normal in 对每个通道单独进行, 或所有通道同时进行.
"""
data_normalized = np.zeros(data.shape, dtype=np.float32)
if isinstance(data, np.ndarray):
data_shape = tuple(list(data.shape))
elif isinstance(data, (list, tuple)):
assert len(data) > 0 and isinstance(data[0], np.ndarray)
data_shape = [len(data)] + list(data[0].shape)
else:
raise TypeError("Data has to be either a numpy array or a list")
if per_channel and isinstance(mean, float) and isinstance(std, float):
mean = [mean] * data_shape[1]
std = [std] * data_shape[1]
elif per_channel and isinstance(mean, (tuple, list, np.ndarray)):
assert len(mean) == data_shape[1]
elif per_channel and isinstance(std, (tuple, list, np.ndarray)):
assert len(std) == data_shape[1]
for b in range(data_shape[0]):
if per_channel:
for c in range(data_shape[1]):
data_normalized[b][c] = (data[b][c] - mean[c]) / std[c]
else:
data_normalized[b] = (data[b] - mean) / std
return data_normalized
def cut_off_outliers(data, percentile_lower=0.2, percentile_upper=99.8, per_channel=False):
r"""截断部分像素值, 可以截断不需要的目标干扰
@Args:
百分比范围(percentile_lower, percentile_upper)
"""
for b in range(len(data)):
if per_channel:
for c in range(data.shape[1]):
cut_off_lower = np.percentile(data[b, c], percentile_lower)
cut_off_upper = np.percentile(data[b, c], percentile_upper)
data[b, c][data[b, c] < cut_off_lower] = cut_off_lower
data[b, c][data[b, c] > cut_off_upper] = cut_off_upper
pass
pass
else:
cut_off_lower = np.percentile(data[b], percentile_lower)
cut_off_upper = np.percentile(data[b], percentile_upper)
data[b][data[b] < cut_off_lower] = cut_off_lower
data[b][data[b] > cut_off_upper] = cut_off_upper
pass
pass
return data
def test():
import os
import numpy as np
import SimpleITK as sitk
def plot_3view(image_norm):
b, c, z, y, x = image_norm.shape
import matplotlib.pyplot as plt
plt.figure(figsize=(10,10))
plt.subplot(131)
plt.imshow(image_norm[0,0, z//2, ::, ::]) # x,y,z
plt.subplot(132)
plt.imshow(image_norm[0,0, ::, y//2, ::]) # x,y,z
plt.subplot(133)
plt.imshow(image_norm[0,0, ::, ::, x//2]) # x,y,z
plt.show()
pass
mhd_path = r'xxx.mhd'
sitk_image = sitk.ReadImage(mhd_path) # xyz
x,y,z = sitk_image.GetSize()
data = sitk.GetArrayFromImage(sitk_image) # zyx
data.shape # zyx
data = data[np.newaxis, np.newaxis] # nczyx
# 可视化归一化 1
image_norm = range_normalization(data, per_channel=True)
print(image_norm.min(), image_norm.max())
plot_3view(image_norm)
# 可视化归一化 2
image_norm = zero_mean_unit_variance_normalization(data, False)
print(image_norm.min(), image_norm.max())
plot_3view(image_norm)
image_norm = zero_mean_unit_variance_normalization(data, True)
print(image_norm.min(), image_norm.max())
plot_3view(image_norm)
# 可视化像素截断 3
image_norm = mean_std_normalization(data, mean=43., std=52., per_channel=True)
print(image_norm.min(), image_norm.max())
plot_3view(image_norm)
# 可视化像素截断 4
image_norm = cut_off_outliers(data.copy(), percentile_lower=0.2, percentile_upper=95, per_channel=False)
plot_3view(data)
plot_3view(image_norm)
if __name__=="__main__":
test()
边栏推荐
- PHP中使用XML-RPC构造Web Service简单入门
- KMP Media Group South Africa implemented a DMS (Document Management System) to digitize the process, employees can again focus on their actual tasks, providing efficiency
- Implementation of FIR filter based on FPGA (1) - using fir1 function design
- php文件上传下载(存放文件二进制到数据库)
- LeetCode简单题之统计星号
- R语言ggplot2可视化:使用ggpubr包的ggdonutchart函数可视化甜甜圈图(donut chart)、为甜甜圈图添加自定义标签(包含文本内容以及数值百分比)、lab.font参数设置标
- idea中项目呈现树形结构
- 全网最全的PADS 9.5安装教程与资源包
- 【Personal Summary】2022.8.7 Week End
- 清华|GLM-130B:一个开放的双语预训练模型
猜你喜欢

难产的“第一股”:中式快餐之困

化工行业数字化供应链系统:赋能化工企业高质量发展,促进上下游协同
Implement a customized pin code input control

Program Environment and Preprocessing

keil5——安装教程附资源包

哈佛大学砸场子:DALL-E 2只是「粘合怪」,生成正确率只有22%
【Redis】redis安装与客户端redis-cli的使用(批量操作)

机器学习+深度学习笔记(持续更新~)
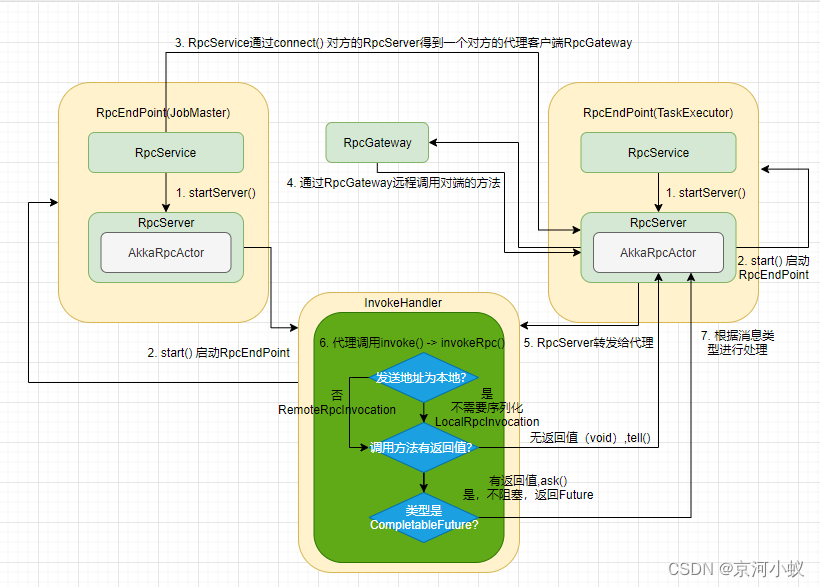
Flink1.15 组件RPC通信过程概览图
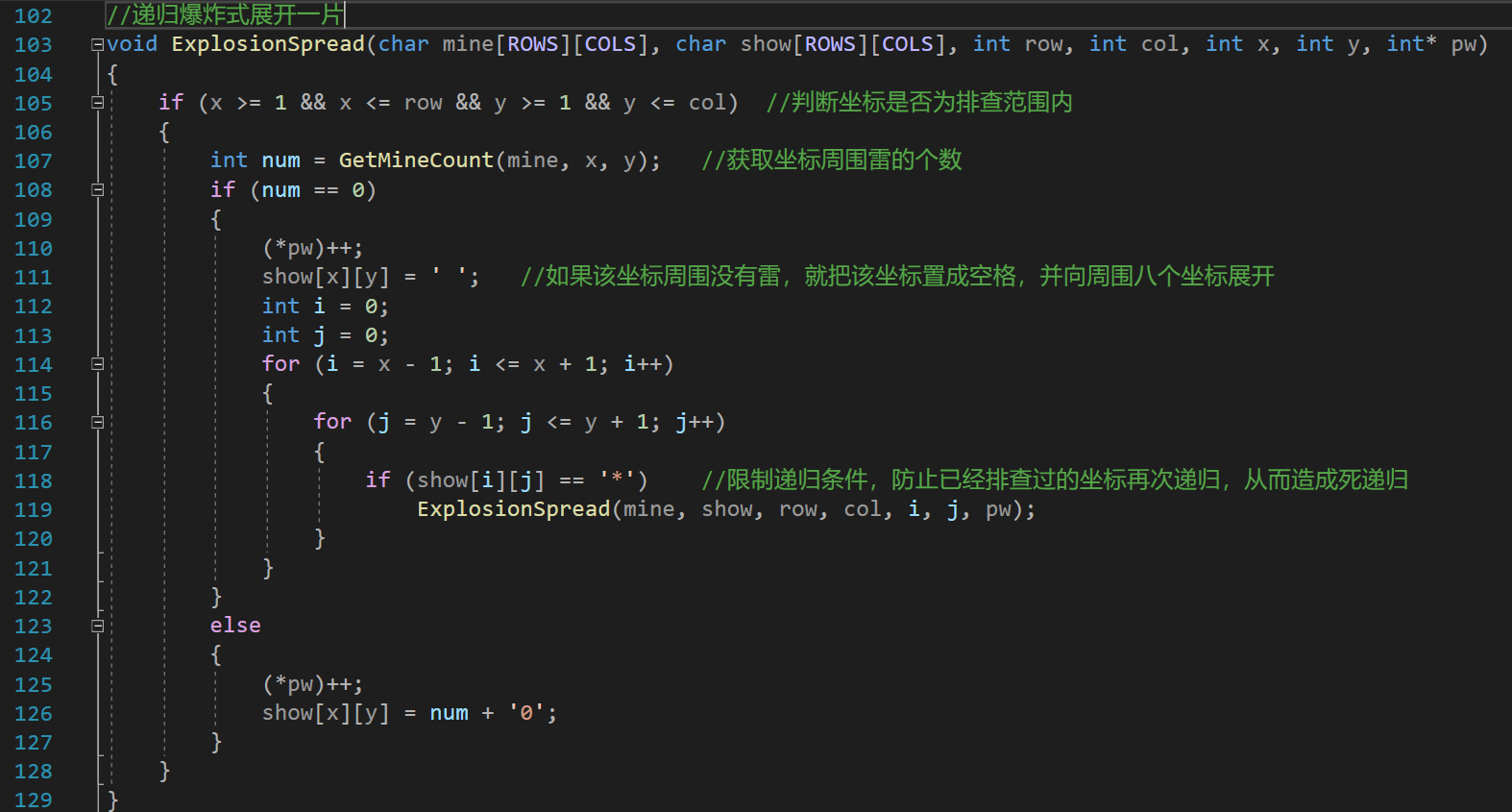
C language small project - complete code of minesweeper game (recursive expansion + selection mark)
随机推荐
今日睡眠质量记录83分
《预训练周刊》第56期:长文本理解、即时问答、掩码自监督
又一个千亿市场,冰淇淋也成了创新试验田
直接选择排序
poj3744 Scout YYF I
Time to update your tech arsenal in 2020: Asgi vs Wsgi (FastAPI vs Flask)
(7) FlinkSQL kafka data written to the mysql way 2
[Redis] Redis installation and use of client redis-cli (batch operation)
HackTheBox | Previse
Server Configuration - Install Redis on Linux System
Qt的简易日志库实现及封装
Photoshop插件-charIDToTypeID-PIStringTerminology.h-不同值的解释及参考-脚本开发-PS插件
MySQL:索引(1)原理与底层结构
R语言ggpubr包的ggsummarystats函数可视化分面箱图(通过ggfunc参数和facet.by参数设置)、添加描述性统计结果表格、palette参数配置不同水平可视化图像和统计值的颜色
全网最全的AItium Designer 16下载资源与安装步骤
MapStruct入门使用
C language small project -- address book (static version + dynamic version + file version)
译文推荐|深入解析 BookKeeper 协议模型与验证
数据解析(XPath、BeautifulSoup、正则表达式、pyquery)
PHP中使用XML-RPC构造Web Service简单入门