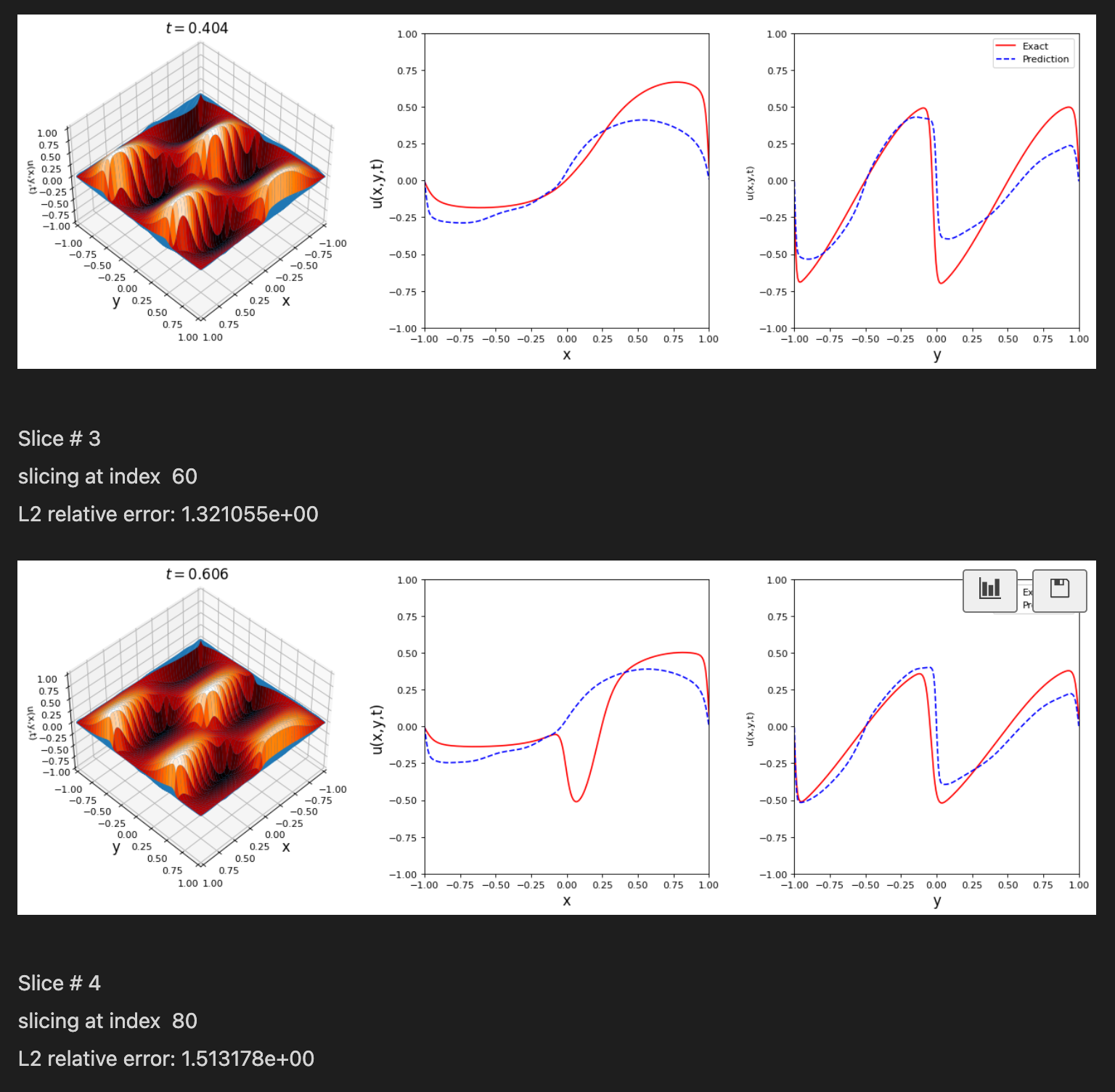
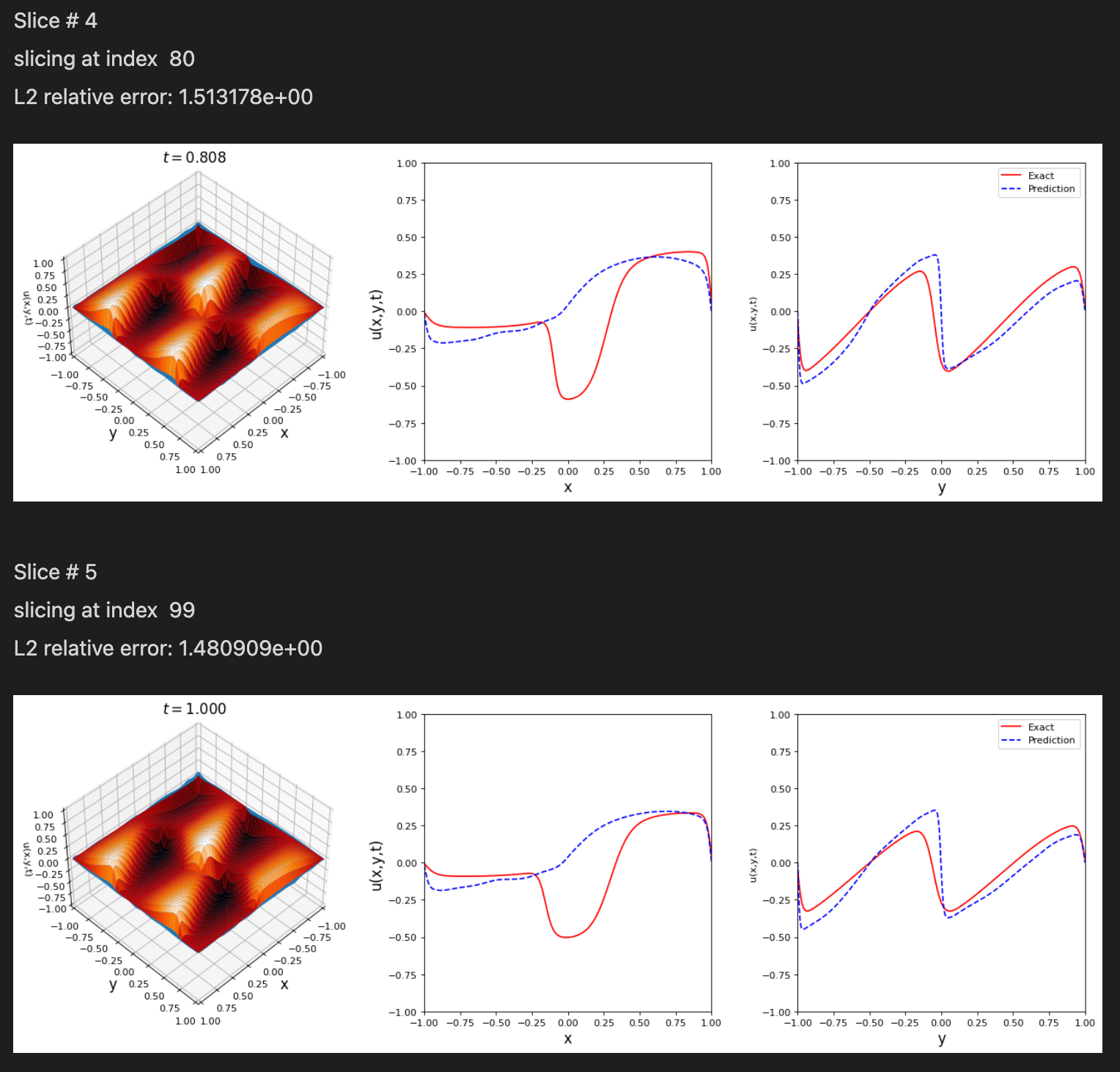
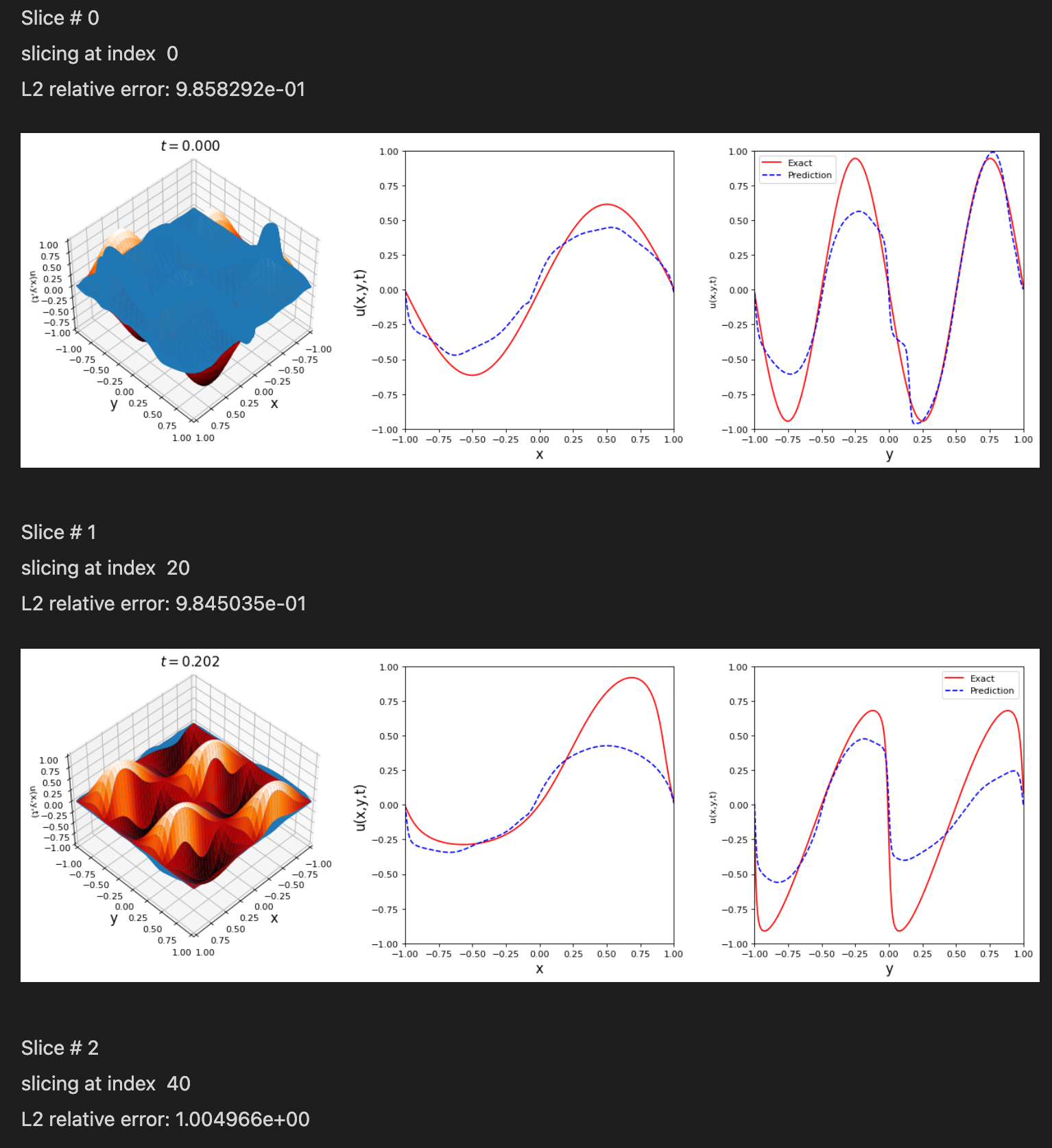
Notice: Support for Python 3.6 will be dropped in v.0.2.1, please plan accordingly!
Efficient and Scalable Physics-Informed Deep Learning
Collocation-based PINN PDE solvers for prediction and discovery methods on top of Tensorflow 2.X for multi-worker distributed computing.
Use TensorDiffEq if you require:
- A meshless PINN solver that can distribute over multiple workers (GPUs) for forward problems (inference) and inverse problems (discovery)
- Scalable domains - Iterated solver construction allows for N-D spatio-temporal support
- support for N-D spatial domains with no time element is included
- Self-Adaptive Collocation methods for forward and inverse PINNs
- Intuitive user interface allowing for explicit definitions of variable domains, boundary conditions, initial conditions, and strong-form PDEs
What makes TensorDiffEq different?
-
Completely open-source
-
Self-Adaptive Solvers for forward and inverse problems, leading to increased accuracy of the solution and stability in training, resulting in less overall training time
-
Multi-GPU distributed training for large or fine-grain spatio-temporal domains
-
Built on top of Tensorflow 2.0 for increased support in new functionality exclusive to recent TF releases, such as XLA support, autograph for efficent graph-building, and grappler support for graph optimization* - with no chance of the source code being sunset in a further Tensorflow version release
-
Intuitive interface - defining domains, BCs, ICs, and strong-form PDEs in "plain english"
*In development
If you use TensorDiffEq in your work, please cite it via:
@article{mcclenny2021tensordiffeq,
title={TensorDiffEq: Scalable Multi-GPU Forward and Inverse Solvers for Physics Informed Neural Networks},
author={McClenny, Levi D and Haile, Mulugeta A and Braga-Neto, Ulisses M},
journal={arXiv preprint arXiv:2103.16034},
year={2021}
}
Thanks to our additional contributors:
@marcelodallaqua, @ragusa, @emiliocoutinho