当前位置:网站首页>A new method for evaluating the quality of metagenome assembly - magista
A new method for evaluating the quality of metagenome assembly - magista
2022-04-23 04:11:00 【Gu he Niubo】
Gu He health

Despite the diversity of microbial groups on earth , but Only a small part Got it Culture and effective naming . Because most bacteria cannot... Under very specific conditions Culture, isolation and identification .
In the past decade , Metagenomic research The importance of has been highlighted , Because it can evaluate bacterial gene bank and find out what current laboratory culture technology can't master New bacterial genome . These data are for expand Our understanding of microbial diversity on earth is crucial .
Because the metagenome sequencing data is from Multiple species and bacteria Plant DNA The sequence consists of fragments , There are usually thousands from different Life field , Therefore, the main challenge of such analysis is to correctly identify each DNA Sequence fragment The real source . Unfortunately , These steps It's easy to make mistakes , Therefore, the results must be strictly reviewed , To avoid publishing incomplete and low-quality genomes .
lately , Belgian researchers have newly developed MAGISTA, This is an assessment of metagenomes A new method of assembly quality , Estimation based on random forest method MAGs Integrity and contamination of , Solves current problems based on Some defects of the method of reference gene are often ignored .
MAGISTA It's based on the metagenome bins Inside Of contig fragment Non aligned distance distribution between , Instead of a set of reference genes . This method utilizes data from the whole bin Information about . In order to properly evaluate this method , And explain the disadvantages of reference based tools , lately , Belgian researchers have built a highly complex DNA Simulated community , from 227 It consists of three bacterial strains , And have different degrees of similarity .
Fang Law
Training set to (HC227) since 227 A bacterial strain , The test data set consists of five publicly available Short reading (short reads) Subsets make up , There are four relatively low complexity genomes DNA Simulated community reads. See the figure below for details .
Complexity List the number of indicator bacteria ;Assembly tool The list shows the software used for assembly ;Binning method The list shows the tools used to divide boxes ;Binning parameters The list shows the used for Evaluate the quality of crating Indicators of ,comp by Completeness ,cov by coverage .

MAGISTA computational procedure :
Input binning After each bins
-●-
The first 1 Step : Select the appropriate fragment size and distance calculation method
-●-
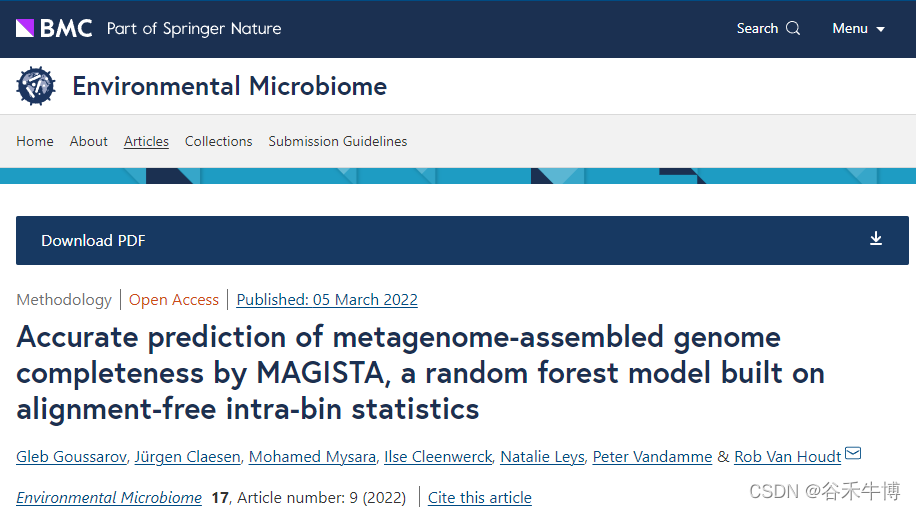
First, put each bin Each of the contig Split into fixed length segments , And then use Four different methods ( namely PaSiT4、MMZ3、MMZ4 and Freq4) Calculate a bin All distances between clips in . For each method , All selected a specific fragment length , In order to produce different characteristic distributions for different organisms .
Of each method Final fragment length Your choice is through Different methods of analysis and integration Decisive , The method is shown in the figure below . At least two genomes in each group's design come from the same family , The two genomes come from the same order but from different families . These genomes are artificially divided into fragments of the required length , And calculate the target feature for each segment .
For each group of five genomes , All fragments were mixed and principal component analysis was performed according to their characteristics (PCA), Then proceed Quadratic discrimination analysis , Used to generate classifiers , The aim is to distinguish the two most overlapping genomes in each group . Of the classifier The accuracy is averaged , The results are used to select the final combination of method and fragment length .
-●-
The first 2 Step : Selection of characteristic variables in the model
-●-
After selecting the fragment length for each method , Use average 、 Standard deviation 、 skewness 、 Kurtosis and median and 2.5%、5%、10%、90%、95% and 97.5% The percentile calculates the distance distribution . Besides , And calculated 1 kb Fragment GC Content distribution . And each bin Size , total 66 Characteristic variables .
-●-
The first 3 Step : model building
-●-
Use R (v 4.0.3) package “RandomForest” Medium “RandomForest” Function and default parameters to train the random forest model . Use at the same time R package lm Then a linear model is established to perform linear regression , Input the characteristic variable value after logarithmic conversion , For cross validation analysis .
Lord want junction fruit
A highly complex genome DNA Simulated community
From 227 The genome of a bacterial strain DNA form , These strains belong to 8 A door (Actinobacteria, Bacteroidetes,Deinococcus-Thermus, Firmicutes,Fusobacteria,Planctomycetes, Proteobacteria and Verrucomicrobia),18 class ,47 Objective ,85 Families, ,175 Belong to ,197 Kind of .

edit
The above figure shows the genome size and size of bacterial strains in the simulated community GC content ( from 26.3% To 73.4%) Scatter plot ;

edit
The figure shows the relationship between species in the training set and the test set . The red line indicates the species that exist in the training set , The gray line indicates the bacteria in the training set . Different colors in the ring chart represent different classification levels . In the legend, there are bacteria in the training set * Mark , The phylum present in archaea is marked with dark gray bands .
CheckM Based on single copy markers (SCMG) To assess the bin Defects in quality
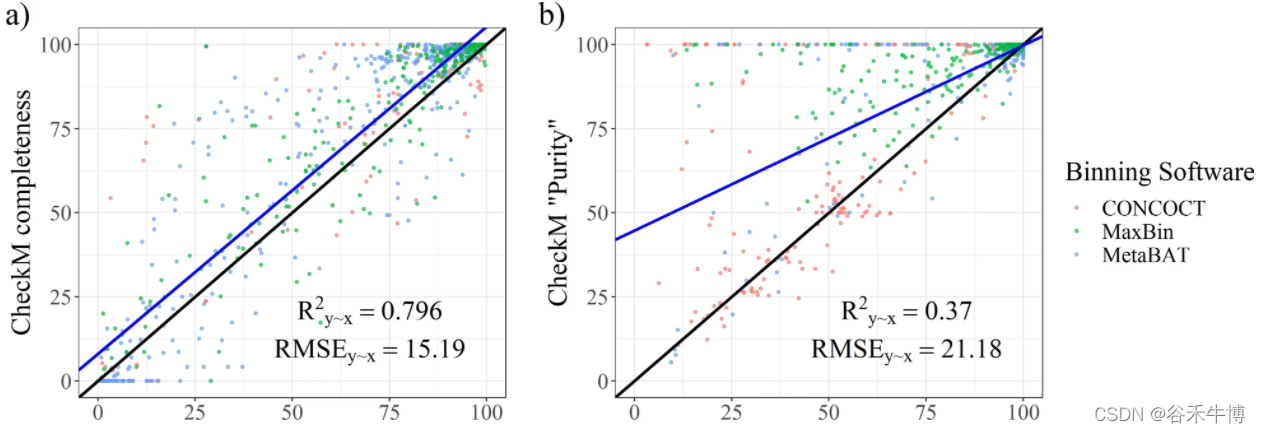
chart a and b They are from CheckM Integrity index and pollution degree of output in . Use R^2y∼x( Explain the percentage of variance ),RMSE( Root mean square error relative to actual value ) Two parameter evaluation results . The result shows CheckM Overestimate bin The quality of the . Many contaminated bins Predicted to be close to uncontaminated .
Use MAGISTA Analyze the bins

First, the optimal fragment size is selected to calculate the distance distribution , As shown in the figure above , Considering 1、5、10、20、30、40、50、75 and 100 kb Fragments of , Finally, the fragment size shown in bold is selected .

The graph is concont、MetaBAT and MaxBin Produced bins Integrity and contamination information .
Because generating such a data set through simulation can not accurately represent the real results , So we used binning Software Result , Provides a set of Different quality real bins. The integrity and uncontaminated degree of the training data set are in 90% above .

Finally, the model construction , establish Prediction model of integrity and pollution degree . The model is evaluated , As shown in the figure . Respectively for CheckM、MAGISTA and MAGISTIC Its performance is tested .CheckM It is now a mainstream evaluation bin Quality tools .MAGISTIC It's a combination of CheckM and MAGISTA Tools for . Use scores that explain variance (R2y∼x) And root mean square error (RMSE) As an indicator of performance . Prediction of integrity ,MAGISTA be better than CheckM. Prediction of pollution degree ,MAGISTA Better than CheckM.
junction On
Researchers have developed a new method for predicting highly complex metagenome assembly genomes bin The quality of the method ,MAGISTA. Is based on SCMG An equally good alternative to the low complexity metagenomic approach . except MAGISTA outside , Also by combining CheckM Result , Use MAGISTIC Generated a more accurate prediction .
The researchers pointed out in the article MAGISTA and CheckM Are not accurate enough to be considered reliable .MAGISTIC Produced a better than MAGISTA Better results .
In additional analysis , Divide the test set into two subsets , From reality and simulation reads From bins, Analyze this again , The result shows ,CheckM about “ real ” Subsets perform well ( But compared with MAGISTA and MAGISTIC Still poor ), about “ simulation ” Subsets perform poorly . and MAGISTIC comparison MAGISTA It would be more accurate . however The article does not elaborate MAGISTIC workflow .
See the author in github Software description disclosed on , The address is as follows . However, the contents of the output file are not explained and given . Personally, I think it may not be very mature .
reference :
Goussarov G, Claesen J, Mysara M, Cleenwerck I, Leys N, Vandamme P, Van Houdt R. Accurate prediction of metagenome-assembled genome completeness by MAGISTA, a random forest model built on alignment-free intra-bin statistics. Environ Microbiome. 2022 Mar 5;17(1):9. doi: 10.1186/s40793-022-00403-7. PMID: 35248155; PMCID: PMC8898458.
版权声明
本文为[Gu he Niubo]所创,转载请带上原文链接,感谢
https://yzsam.com/2022/04/202204220600369475.html
边栏推荐
- STM32上μC/Shell移植与应用
- 一个函数秒杀2Sum 3Sum 4Sum问题
- 作为一名码农,女友比自己更能码是一种什么体验?
- Qt程序集成EasyPlayer-RTSP流媒体播放器出现画面闪烁是什么原因?
- 【测绘程序设计】坐标方位角推算神器(C#版)
- [AI vision · quick review of robot papers today, issue 30] Thu, 14 APR 2022
- What is software acceptance testing? What are the benefits of acceptance testing conducted by third-party software testing institutions?
- Single chip microcomputer serial port data processing (1) -- serial port interrupt sending data
- [mapping program design] coordinate inverse artifact v1 0 (with C / C / VB source program)
- Installation and configuration of clion under win10
猜你喜欢
【测绘程序设计】坐标方位角推算神器(C#版)

Why recommend you to study embedded

创下国产手机在海外市场销量最高纪录的小米,重新关注国内市场

Understand the gut organ axis, good gut and good health
STM32单片机ADC规则组多通道转换-DMA模式

Common string processing functions in C language

为什么推荐你学嵌入式
Photoshop installation under win10

Xiaohongshu was exposed to layoffs of 20% as a whole, and the internal volume among large factories was also very serious

Qtspim manual - Chinese Translation
随机推荐
【BIM+GIS】ArcGIS Pro2. 8 how to open Revit model, Bim and GIS integration?
Unipolar NRZ code, bipolar NRZ code, 2ASK, 2FSK, 2PSK, 2DPSK and MATLAB simulation
AI CC 2019 installation tutorial under win10 (super detailed - small white version)
Express middleware ① (use of Middleware)
QT program integration easyplayer RTSP streaming media player screen flicker what is the reason?
Qt程序集成EasyPlayer-RTSP流媒体播放器出现画面闪烁是什么原因?
【Echart】echart 入門
[mathematical modeling] my mathematical memory
Qtspim manual - Chinese Translation
The whole process of connecting the newly created unbutu system virtual machine with xshell and xftp
列表、元组、字典和集合的区别
What if win10 doesn't have a local group policy?
MYSQL去重方法汇总
[Li Hongyi 2022 machine learning spring] hw6_ Gan (don't understand...)
[AI vision · quick review of NLP natural language processing papers today, issue 28] wed, 1 Dec 2021
单片机串口数据处理(1)——串口中断发送数据
上海航芯技术分享 | ACM32 MCU安全特性概述
基于PHP的代步工具购物商城
中国移动日赚2.85亿很高?其实是5G难带来更多利润,那么钱去哪里了?
Overview of knowledge map (II)