当前位置:网站首页>Elmo (bilstm-crf + Elmo) (conll-2003 named entity recognition NER)
Elmo (bilstm-crf + Elmo) (conll-2003 named entity recognition NER)
2022-04-23 13:54:00 【Shallow singing under the hedge】
List of articles
- elmo(BiLSTM-CRF+elmo)(Conll-2003 Named entity recognition NER)
- One 、 File directory
- Two 、 Corpus
- 3、 ... and 、 Data processing (bulid_data.py)(data_utils.py)
- Four 、NERModel Model (ner_model.py)
- 5、 ... and 、BiLSTM-CRF+ELMO Model training process (ner_learner.py)
- 6、 ... and 、 Calculation loss value (CRF)
- 7、 ... and 、 Training (train.py)
- 8、 ... and 、 test (test.py)
- experimental result
elmo(BiLSTM-CRF+elmo)(Conll-2003 Named entity recognition NER)
Bidirectional laguage models:

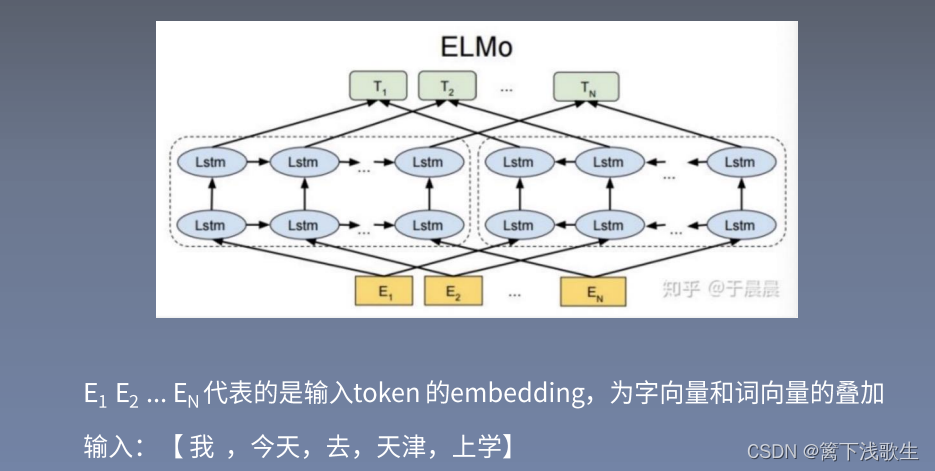

elmo:


elmo Downstream tasks :


One 、 File directory


Two 、 Corpus
CoNLL 2003 NER :

The first column of the dataset is the word , The second column is part of speech , The third column is the syntax , The fourth column is the entity label . stay NER Tasks , Only care about the first and fourth Columns .
3、 ... and 、 Data processing (bulid_data.py)(data_utils.py)
bulid_data.py—— Data processing
from model.config import Config
from model.data_utils import CoNLLDataset, get_vocabs, UNK, NUM, \
get_glove_vocab, write_vocab, load_vocab, get_char_vocab, \
export_trimmed_glove_vectors, get_processing_word
def main():
"""Procedure to build data You MUST RUN this procedure. It iterates over the whole dataset (train, dev and test) and extract the vocabularies in terms of words, tags, and characters. Having built the vocabularies it writes them in a file. The writing of vocabulary in a file assigns an id (the line #) to each word. It then extract the relevant GloVe vectors and stores them in a np array such that the i-th entry corresponds to the i-th word in the vocabulary. Args: config: (instance of Config) has attributes like hyper-params... """
# 1. get config and processing of words
config = Config(load=False)
#2. Get processing word generator
# Get pre training words embedding
processing_word = get_processing_word(lowercase=True)
# 3. Generators
# get dev,test,train Of word and target
dev = CoNLLDataset(config.filename_dev, processing_word)
test = CoNLLDataset(config.filename_test, processing_word)
train = CoNLLDataset(config.filename_train, processing_word)
# 4. Build Word and Tag vocab
# establish word and target The thesaurus of
vocab_words, vocab_tags = get_vocabs([train, dev, test])
# Handle glove: Jianli glove Thesaurus
vocab_glove = get_glove_vocab(config.filename_glove)
# 5. Get a vocab set for words in both vocab_words and vocab_glove
# vocab For existing vocab_words Also exist vocab_glove The word
vocab = vocab_words & vocab_glove
# vocab Add UNK,NUM
vocab.add(UNK)
vocab.add(NUM)
# 6. Save vocab
# preservation words.txt and tags.txt
write_vocab(vocab, config.filename_words)
write_vocab(vocab_tags, config.filename_tags)
# 7. Trim GloVe Vectors
# download vocab
vocab = load_vocab(config.filename_words)
# Get the word vector corresponding to the word
export_trimmed_glove_vectors(vocab, config.filename_glove,
config.filename_trimmed, config.dim_word)
# Build and save char vocab
# get train Of word and target
train = CoNLLDataset(config.filename_train)
# Get training focus word Of char
vocab_chars = get_char_vocab(train)
# preservation chars.txt
write_vocab(vocab_chars, config.filename_chars)
if __name__ == "__main__":
main()
data_utils.py—— Various functions of data processing
" Data utils from https://github.com/guillaumegenthial/sequence_tagging "
import numpy as np
import torch
import os
# shared global variables to be imported from model also
UNK = "$UNK$"
NUM = "$NUM$"
NONE = "O"
# special error message
class MyIOError(Exception):
def __init__(self, filename):
# custom error message
message = """ ERROR: Unable to locate file {}. FIX: Have you tried running python build_data.py first? This will build vocab file from your train, test and dev sets and trimm your word vectors. """.format(filename)
super(MyIOError, self).__init__(message)
# Training data preprocessing
# Function call :train = CoNLLDataset(config.filename_train, processing_word)
class CoNLLDataset(object):
"""Class that iterates over CoNLL Dataset __iter__ method yields a tuple (words, tags) words: list of raw words tags: list of raw tags If processing_word and processing_tag are not None, optional preprocessing is appplied Example: ```python data = CoNLLDataset(filename) for sentence, tags in data: pass ``` """
def __init__(self, filename, processing_word=None, processing_tag=None,
max_iter=None, use_crf=True):
""" Args: filename: path to the file processing_words: (optional) function that takes a word as input processing_tags: (optional) function that takes a tag as input max_iter: (optional) max number of sentences to yield """
self.filename = filename
# Preprocessing words
self.processing_word = processing_word
# Handle labels
self.processing_tag = processing_tag
# The number of iterations
self.max_iter = max_iter
# Whether to use crf
self.use_crf = use_crf
self.length = None
def __iter__(self):
niter = 0
with open(self.filename) as f:
words, tags = [], []
for line in f:
line = line.strip()
# End with a sentence
if (len(line) == 0 or line.startswith("-DOCSTART-")):
if len(words) != 0:
niter += 1
if self.max_iter is not None and niter > self.max_iter:
break
# preservation words, tags
yield words, tags
# Empty words, tags
words, tags = [], []
else:
ls = line.split(' ')
# Get the first column ( word ) And the fourth column ( label )
word, tag = ls[0],ls[-1]
if self.processing_word is not None:
word = self.processing_word(word)
if self.processing_tag is not None:
if self.use_crf:
tag = self.processing_tag(tag)
words += [word]
tags += [tag]
def __len__(self):
"""Iterates once over the corpus to set and store length"""
if self.length is None:
self.length = 0
for _ in self:
self.length += 1
return self.length
# Get words and tags
# Function call :vocab_words, vocab_tags = get_vocabs([train, dev, test])
def get_vocabs(datasets):
"""Build vocabulary from an iterable of datasets objects Args: datasets: a list of dataset objects Returns: a set of all the words in the dataset """
print("Building vocab...")
vocab_words = set()
vocab_tags = set()
for dataset in datasets:
for words, tags in dataset:
vocab_words.update(words)
vocab_tags.update(tags)
print("- done. {} tokens".format(len(vocab_words)))
return vocab_words, vocab_tags
# Get training focused char
# Function transfer :vocab_chars = get_char_vocab(train)
def get_char_vocab(dataset):
"""Build char vocabulary from an iterable of datasets objects Args: dataset: a iterator yielding tuples (sentence, tags) Returns: a set of all the characters in the dataset """
print("Building char vocab...")
vocab_char = set()
for words, _ in dataset:
for word in words:
vocab_char.update(word)
print("- done. {} tokens".format(len(vocab_char)))
return vocab_char
# Handle glove: Jianli glove Thesaurus
# Function call :vocab_glove = get_glove_vocab(config.filename_glove)
def get_glove_vocab(filename):
"""Load vocab from file Args: filename: path to the glove vectors Returns: vocab: set() of strings """
print("Building vocab...")
vocab = set()
with open(filename, encoding="utf8") as f:
for line in f:
word = line.strip().split(' ')[0]
#glove The first word of is stored in vocab in
vocab.add(word)
print("- done. {} tokens".format(len(vocab)))
return vocab
def write_vocab(vocab, filename):
"""Writes a vocab to a file Writes one word per line. Args: vocab: iterable that yields word filename: path to vocab file Returns: write a word per line """
print("Writing vocab...")
with open(filename, "w") as f:
for i, word in enumerate(vocab):
if i != len(vocab) - 1:
f.write("{}\n".format(word))
else:
f.write(word)
print("- done. {} tokens".format(len(vocab)))
# Get a dictionary word and id
# Function call : self.vocab_words = load_vocab(self.filename_words)
def load_vocab(filename):
"""Loads vocab from a file Args: filename: (string) the format of the file must be one word per line. Returns: d: dict[word] = index """
try:
d = dict()
with open(filename) as f:
for idx, word in enumerate(f):
word = word.strip()
d[word] = idx
except IOError:
raise MyIOError(filename)
return d
# Get the word vector corresponding to the word
# Function call :export_trimmed_glove_vectors(vocab, config.filename_glove,config.filename_trimmed, config.dim_word)
def export_trimmed_glove_vectors(vocab, glove_filename, trimmed_filename, dim):
"""Saves glove vectors in numpy array Args: vocab: dictionary vocab[word] = index glove_filename: a path to a glove file trimmed_filename: a path where to store a matrix in npy dim: (int) dimension of embeddings """
embeddings = np.zeros([len(vocab), dim])
with open(glove_filename, encoding="utf8") as f:
for line in f:
line = line.strip().split(' ')
word = line[0]
embedding = [float(x) for x in line[1:]]
if word in vocab:
word_idx = vocab[word]
embeddings[word_idx] = np.asarray(embedding)
np.savez_compressed(trimmed_filename, embeddings=embeddings)
def get_trimmed_glove_vectors(filename):
""" Args: filename: path to the npz file Returns: matrix of embeddings (np array) """
try:
with np.load(filename) as data:
return data["embeddings"]
except IOError:
raise MyIOError(filename)
# Get pre training words
# Initialization function call :processing_word = get_processing_word(lowercase=True)
# Function call :self.processing_word = get_processing_word(self.vocab_words,self.vocab_chars, lowercase=True, chars=self.use_chars)
def get_processing_word(vocab_words=None, vocab_chars=None,
lowercase=False, chars=False, allow_unk=True):
"""Return lambda function that transform a word (string) into list, or tuple of (list, id) of int corresponding to the ids of the word and its corresponding characters. Args: vocab: dict[word] = idx Returns: f("cat") = ([12, 4, 32], 12345) = (list of char ids, word id) """
def f(word):
# 0. get chars of words
#chars == False
if vocab_chars is not None and chars == True:
char_ids = []
for char in word:
# ignore chars out of vocabulary
if char in vocab_chars:
char_ids += [vocab_chars[char]]
# 1. preprocess word
# lowercase=True
if lowercase:
word = word.lower()
# If word It's the number. , Turn to special symbols NUM
if word.isdigit():
word = NUM
# 2. get id of word
# vocab_words=None
if vocab_words is not None:
if word in vocab_words:
word = vocab_words[word]
else:
if allow_unk:
word = vocab_words[UNK]
else:
raise Exception("Unknow key is not allowed. Check that "\
"your vocab (tags?) is correct")
# 3. return tuple char ids, word id
if vocab_chars is not None and chars == True:
return char_ids, word
else:
return word
return f
def _pad_sequences(sequences, pad_tok, max_length):
""" Args: sequences: a generator of list or tuple pad_tok: the char to pad with Returns: a list of list where each sublist has same length """
sequence_padded, sequence_length = [], []
for seq in sequences:
seq = list(seq)
seq_ = seq[:max_length] + [pad_tok]*max(max_length - len(seq), 0)
sequence_padded += [seq_]
sequence_length += [min(len(seq), max_length)]
return sequence_padded, sequence_length
def pad_sequences(sequences, pad_tok, nlevels=1):
""" Args: sequences: a generator of list or tuple pad_tok: the char to pad with nlevels: "depth" of padding, for the case where we have characters ids Returns: a list of list where each sublist has same length """
if nlevels == 1:
max_length = max(map(lambda x : len(x), sequences))
sequence_padded, sequence_length = _pad_sequences(sequences,
pad_tok, max_length)
elif nlevels == 2:
max_length_word = max([max(map(lambda x: len(x), seq))
for seq in sequences])
sequence_padded, sequence_length = [], []
for seq in sequences:
# all words are same length now
sp, sl = _pad_sequences(seq, pad_tok, max_length_word)
sequence_padded += [sp]
sequence_length += [sl]
max_length_sentence = max(map(lambda x : len(x), sequences))
sequence_padded, _ = _pad_sequences(sequence_padded,
[pad_tok]*max_length_word, max_length_sentence)
sequence_length, _ = _pad_sequences(sequence_length, 0,
max_length_sentence)
return sequence_padded, sequence_length
def minibatches(data, minibatch_size, use_crf=True):
""" Args: data: generator of (sentence, tags) tuples minibatch_size: (int) Yields: list of tuples """
x_batch, y_batch = [], []
for (x, y) in data:
if len(x_batch) == minibatch_size:
yield x_batch, y_batch
x_batch, y_batch = [], []
if type(x[0]) == tuple:
x = zip(*x)
x_batch += [x]
if use_crf:
y_batch += [y]
else:
if any([x.isdigit() for x in y]):
y_batch.append([int(x) for x in y if x.isdigit()])
else:
y_batch.append([0,0,0,0,0])
if len(x_batch) != 0:
yield x_batch, y_batch
def get_chunk_type(tok, idx_to_tag):
""" Args: tok: id of token, ex 4 idx_to_tag: dictionary {4: "B-PER", ...} Returns: tuple: "B", "PER" """
if isinstance(tok, torch.Tensor): tok = tok.item()
tag_name = idx_to_tag[tok]
tag_class = tag_name.split('-')[0]
tag_type = tag_name.split('-')[-1]
return tag_class, tag_type
def get_chunks(seq, tags):
"""Given a sequence of tags, group entities and their position Args: seq: [4, 4, 0, 0, ...] sequence of labels tags: dict["O"] = 4 Returns: list of (chunk_type, chunk_start, chunk_end) Example: seq = [4, 5, 0, 3] tags = {"B-PER": 4, "I-PER": 5, "B-LOC": 3} result = [("PER", 0, 2), ("LOC", 3, 4)] """
default = tags[NONE]
idx_to_tag = {
idx: tag for tag, idx in tags.items()}
chunks = []
chunk_type, chunk_start = None, None
for i, tok in enumerate(seq):
# End of a chunk 1
if tok == default and chunk_type is not None:
# Add a chunk.
chunk = (chunk_type, chunk_start, i)
chunks.append(chunk)
chunk_type, chunk_start = None, None
# End of a chunk + start of a chunk!
elif tok != default:
tok_chunk_class, tok_chunk_type = get_chunk_type(tok, idx_to_tag)
if chunk_type is None:
chunk_type, chunk_start = tok_chunk_type, i
elif tok_chunk_type != chunk_type or tok_chunk_class == "B":
chunk = (chunk_type, chunk_start, i)
chunks.append(chunk)
chunk_type, chunk_start = tok_chunk_type, i
else:
pass
# end condition
if chunk_type is not None:
chunk = (chunk_type, chunk_start, len(seq))
chunks.append(chunk)
return chunks
Four 、NERModel Model (ner_model.py)
#from fastai.text import *
from .core import *
# NERModel
# Function call :model = NERModel(config)
class NERModel(nn.Module):
def __init__(self, config):
super().__init__()
self.config = config
self.use_elmo = config.use_elmo
# config.use_elmo=True
if not self.use_elmo:
self.emb = nn.Embedding(self.config.nwords, self.config.dim_word, padding_idx=0)
self.char_embeddings = nn.Embedding(self.config.nchars, self.config.dim_char, padding_idx=0)
self.char_lstm = nn.LSTM(self.config.dim_char, self.config.hidden_size_char, bidirectional=True)
self.dropout = nn.Dropout(p=self.config.dropout)
# BiLSTM 1024*600
self.word_lstm = nn.LSTM(self.config.dim_elmo if self.use_elmo else self.config.dim_word+2*self.config.hidden_size_char,
self.config.hidden_size_lstm, bidirectional=True)#dim_elmo = 1024 , hidden_size_lstm = 300
# Output linear layer :[600,9]
self.linear = LinearClassifier(self.config, layers=[self.config.hidden_size_lstm*2, self.config.ntags], drops=[0.5])#self.ntags = len(self.vocab_tags)=9
def forward(self, input):
# Word_dim = (batch_size x sent_length)
# char_dim = (batch_size x sent_length x word_length)
# self.use_elmo=True
if self.use_elmo:
# [5, 31, 1024]->[31,5,1024]
word_emb = self.dropout(input.transpose(0,1))
else:
word_input, char_input = input[0], input[1]
word_input.transpose_(0,1)
# Word Embedding
word_emb = self.emb(word_input) #shape= S*B*wnh
# Char LSTM
char_emb = self.char_embeddings(char_input.view(-1, char_input.size(2))) #https://stackoverflow.com/questions/47205762/embedding-3d-data-in-pytorch
char_emb = char_emb.view(*char_input.size(), -1) #dim = BxSxWxE
_, (h, c) = self.char_lstm(char_emb.view(-1, char_emb.size(2), char_emb.size(3)).transpose(0,1)) #(num_layers * num_directions, batch, hidden_size) = 2*BS*cnh
char_output = torch.cat((h[0], h[1]), 1) #shape = BS*2cnh
char_output = char_output.view(char_emb.size(0), char_emb.size(1), -1).transpose(0,1) #shape = S*B*2cnh
# Concat char output and word output
word_emb = torch.cat((word_emb, char_output), 2) #shape = S*B*(wnh+2cnh)
word_emb = self.dropout(word_emb)
# Get into BiLSTM [31,5,600]
output, (h, c) = self.word_lstm(word_emb) #shape = S,B,hidden_size_lstm
output = self.dropout(output)
# Enter the linear layer [31,5,9]
output = self.linear(output)
return output #shape = S*B*ntags
# By LinearClassifier() Function call , For linear layers
class LinearBlock(nn.Module):
def __init__(self, ni, nf, drop):
super().__init__()
self.lin = nn.Linear(ni, nf)
self.drop = nn.Dropout(drop)
self.bn = nn.BatchNorm1d(ni)
def forward(self, x):
return self.lin(self.drop(self.bn(x)))
# Output linear layer :[600,9]
# Function call : self.linear = LinearClassifier(self.config, layers=[self.config.hidden_size_lstm*2, self.config.ntags], drops=[0.5])
class LinearClassifier(nn.Module):
def __init__(self, config, layers, drops):
self.config = config
super().__init__()
self.layers = nn.ModuleList([
LinearBlock(layers[i], layers[i + 1], drops[i]) for i in range(len(layers) - 1)])
def forward(self, input):
# [31,5,600]
output = input
sl,bs,_ = output.size()
x = output.view(-1, 2*self.config.hidden_size_lstm)#[155,600]
# [155,9]
for l in self.layers:
l_x = l(x)
x = F.relu(l_x)
return l_x.view(sl, bs, self.config.ntags)# [31,5,9]
5、 ... and 、BiLSTM-CRF+ELMO Model training process (ner_learner.py)
""" Works with pytorch 0.4.0 """
from .core import *
from .data_utils import pad_sequences, minibatches, get_chunks
from .crf import CRF
from .general_utils import Progbar
from torch.optim.lr_scheduler import StepLR
# allennlp Medium elmo
if os.name == "posix": from allennlp.modules.elmo import Elmo, batch_to_ids # AllenNLP is currently only supported on linux
# model = NERModel(config)
# Function call :learn = NERLearner(config, model)
class NERLearner(object):
""" NERLearner class that encapsulates a pytorch nn.Module model and ModelData class Contains methods for training a testing the model """
def __init__(self, config, model):
super().__init__()
self.config = config
self.logger = self.config.logger
self.model = model
self.model_path = config.dir_model
self.use_elmo = config.use_elmo
# structure id_to_tag
self.idx_to_tag = {
idx: tag for tag, idx in
self.config.vocab_tags.items()}
self.criterion = CRF(self.config.ntags)
# Optimizer
self.optimizer = optim.Adam(self.model.parameters())
# use_elmo = True
if self.use_elmo:
options_file = "https://s3-us-west-2.amazonaws.com/allennlp/models/elmo/2x4096_512_2048cnn_2xhighway/elmo_2x4096_512_2048cnn_2xhighway_options.json"
weight_file = "https://s3-us-west-2.amazonaws.com/allennlp/models/elmo/2x4096_512_2048cnn_2xhighway/elmo_2x4096_512_2048cnn_2xhighway_weights.hdf5"
self.elmo = Elmo(options_file, weight_file, 2, dropout=0)
else:
self.load_emb()
if USE_GPU:
self.use_cuda = True
self.logger.info("GPU found.")
self.model = model.cuda()
self.criterion = self.criterion.cuda()
if self.use_elmo:
self.elmo = self.elmo.cuda()
print("Moved elmo to cuda")
else:
self.model = model.cpu()
self.use_cuda = False
self.logger.info("No GPU found.")
def get_model_path(self, name):
return os.path.join(self.model_path,name)+'.h5'
def get_layer_groups(self, do_fc=False):
return children(self.model)
def freeze_to(self, n):
c=self.get_layer_groups()
for l in c:
set_trainable(l, False)
for l in c[n:]:
set_trainable(l, True)
def unfreeze(self):
self.freeze_to(0)
def save(self, name=None):
if not name:
name = self.config.ner_model_path
save_model(self.model, self.get_model_path(name))
self.logger.info(f"Saved model at {
self.get_model_path(name)}")
def load_emb(self):
self.model.emb.weight = nn.Parameter(T(self.config.embeddings))
self.model.emb.weight.requires_grad = False
self.logger.info('Loading pretrained word embeddings')
def load(self, fn=None):
if not fn: fn = self.config.ner_model_path
fn = self.get_model_path(fn)
load_ner_model(self.model, fn, strict=True)
self.logger.info(f"Loaded model from {
fn}")
# according to batch_size Organize data sets
# Function call :nbatches_train, train_generator = self.batch_iter(train, batch_size, return_lengths=True)
def batch_iter(self, train, batch_size, return_lengths=False, shuffle=False, sorter=False):
""" Builds a generator from the given dataloader to be fed into the model Args: train: DataLoader batch_size: size of each batch return_lengths: if True, generator returns a list of sequence lengths for each sample in the batch ie. sequence_lengths = [8,7,4,3] shuffle: if True, shuffles the data for each epoch sorter: if True, uses a sorter to shuffle the data Returns: nbatches: (int) number of batches data_generator: batch generator yielding dict inputs:{'word_ids' : np.array([[padded word_ids in sent1], ...]) 'char_ids': np.array([[[padded char_ids in word1_sent1], ...], [padded char_ids in word1_sent2], ...], ...])} labels: np.array([[padded label_ids in sent1], ...]) sequence_lengths: list([len(sent1), len(sent2), ...]) """
nbatches = (len(train) + batch_size - 1) // batch_size
def data_generator():
while True:
if shuffle: train.shuffle()
elif sorter==True and train.sorter: train.sort()
for i, (words, labels) in enumerate(minibatches(train, batch_size)):
# perform padding of the given data
if self.config.use_chars:
char_ids, word_ids = zip(*words)
word_ids, sequence_lengths = pad_sequences(word_ids, 1)
char_ids, word_lengths = pad_sequences(char_ids, pad_tok=0,
nlevels=2)
else:
word_ids, sequence_lengths = pad_sequences(words, 0)
if self.use_elmo:
word_ids = words
if labels:
labels, _ = pad_sequences(labels, 0)
# if categorical
## labels = [to_categorical(label, num_classes=len(train.tag_itos)) for label in labels]
# build dictionary
inputs = {
"word_ids": np.asarray(word_ids)
}
if self.config.use_chars:
inputs["char_ids"] = np.asarray(char_ids)
if return_lengths:
yield(inputs, np.asarray(labels), sequence_lengths)
else:
yield (inputs, np.asarray(labels))
return (nbatches, data_generator())
def fine_tune(self, train, dev=None):
""" Fine tune the NER model by freezing the pre-trained encoder and training the newly instantiated layers for 1 epochs """
self.logger.info("Fine Tuning Model")
self.fit(train, dev, epochs=1, fine_tune=True)
# Function call :learn.fit(train, dev)
def fit(self, train, dev=None, epochs=None, fine_tune=False):
""" Fits the model to the training dataset and evaluates on the validation set. Saves the model to disk """
if not epochs:
epochs = self.config.nepochs
# batch_size = 5
batch_size = self.config.batch_size
# according to batch_size Organize training data sets
nbatches_train, train_generator = self.batch_iter(train, batch_size,
return_lengths=True)
# according to batch_size Collate validation data sets
if dev:
nbatches_dev, dev_generator = self.batch_iter(dev, batch_size,
return_lengths=True)
# Optimizer
scheduler = StepLR(self.optimizer, step_size=1, gamma=self.config.lr_decay)
if not fine_tune: self.logger.info("Training Model")
f1s = []
for epoch in range(epochs):
scheduler.step()
# Put... In the sentence word Generate elmo Medium id, Get into NERModel Training in the model
self.train(epoch, nbatches_train, train_generator, fine_tune=fine_tune)
if dev:
f1 = self.test(nbatches_dev, dev_generator, fine_tune=fine_tune)
# Early stopping
if len(f1s) > 0:
if f1 < max(f1s[max(-self.config.nepoch_no_imprv, -len(f1s)):]): #if sum([f1 > f1s[max(-i, -len(f1s))] for i in range(1,self.config.nepoch_no_imprv+1)]) == 0:
print("No improvement in the last 3 epochs. Stopping training")
break
else:
f1s.append(f1)
if fine_tune:
self.save(self.config.ner_ft_path)
else :
self.save(self.config.ner_model_path)
# Put... In the sentence word Generate elmo Medium id, Get into NERModel Training in the model
# Function call ; self.train(epoch, nbatches_train, train_generator, fine_tune=fine_tune)
def train(self, epoch, nbatches_train, train_generator, fine_tune=False):
self.logger.info('\nEpoch: %d' % epoch)
self.model.train()
if not self.use_elmo: self.model.emb.weight.requires_grad = False
train_loss = 0
correct = 0
total = 0
total_step = None
# Print progress bar
prog = Progbar(target=nbatches_train)
# inputs:batch input data ,targets: Data corresponds to target,
for batch_idx, (inputs, targets, sequence_lengths) in enumerate(train_generator):
if batch_idx == nbatches_train: break
if inputs['word_ids'].shape[0] == 1:
self.logger.info('Skipping batch of size=1')
continue
total_step = batch_idx
targets = T(targets, cuda=self.use_cuda).transpose(0,1).contiguous()
self.optimizer.zero_grad()
# self.use_elmo=True
if self.use_elmo:
sentences = inputs['word_ids']#list(['EU', 'rejects', 'German', 'call', 'to', 'boycott', 'British', 'lamb', '.']
# Put... In the sentence word Generate elmo Medium id
character_ids = batch_to_ids(sentences)
if self.use_cuda:
character_ids = character_ids.cuda()
# Send the sentence to elmo in
embeddings = self.elmo(character_ids)
# get emlo First floor embedding[5,31,1024]
word_input = embeddings['elmo_representations'][0]
# word_input(batch_size x sent_length x dim_elmo):[5,31,1024] , targets:[31,5]
word_input, targets = Variable(word_input, requires_grad=False), \
Variable(targets)
inputs = (word_input)
else:
word_input = T(inputs['word_ids'], cuda=self.use_cuda)
char_input = T(inputs['char_ids'], cuda=self.use_cuda)
word_input, char_input, targets = Variable(word_input, requires_grad=False), \
Variable(char_input, requires_grad=False),\
Variable(targets)
inputs = (word_input, char_input)
# Send to NERModel In the model , Output is [31,5,9]
outputs = self.model(inputs)
# Create mask
if self.use_elmo:
mask = Variable(embeddings['mask'].transpose(0,1))
if self.use_cuda:
mask = mask.cuda()
else:
mask = create_mask(sequence_lengths, targets, cuda=self.use_cuda)
# Get CRF Loss
# CRF Loss function
loss = -1*self.criterion(outputs, targets, mask=mask)
loss.backward()
self.optimizer.step()
# Callbacks
train_loss += loss.item()
# Find the best path in reverse
predictions = self.criterion.decode(outputs, mask=mask)
masked_targets = mask_targets(targets, sequence_lengths)
t_ = mask.type(torch.LongTensor).sum().item()
total += t_
c_ = sum([1 if p[i] == mt[i] else 0 for p, mt in zip(predictions, masked_targets) for i in range(len(p))])
correct += c_
prog.update(batch_idx + 1, values=[("train loss", loss.item())], exact=[("Accuracy", 100*c_/t_)])
self.logger.info("Train Loss: %.3f, Train Accuracy: %.3f%% (%d/%d)" %(train_loss/(total_step+1), 100.*correct/total, correct, total) )
def test(self, nbatches_val, val_generator, fine_tune=False):
self.model.eval()
accs = []
test_loss = 0
correct_preds = 0
total_correct = 0
total_preds = 0
total_step = None
for batch_idx, (inputs, targets, sequence_lengths) in enumerate(val_generator):
if batch_idx == nbatches_val: break
if inputs['word_ids'].shape[0] == 1:
self.logger.info('Skipping batch of size=1')
continue
total_step = batch_idx
targets = T(targets, cuda=self.use_cuda).transpose(0,1).contiguous()
if self.use_elmo:
sentences = inputs['word_ids']
character_ids = batch_to_ids(sentences)
if self.use_cuda:
character_ids = character_ids.cuda()
embeddings = self.elmo(character_ids)
word_input = embeddings['elmo_representations'][1]
word_input, targets = Variable(word_input, requires_grad=False), \
Variable(targets)
inputs = (word_input)
else:
word_input = T(inputs['word_ids'], cuda=self.use_cuda)
char_input = T(inputs['char_ids'], cuda=self.use_cuda)
word_input, char_input, targets = Variable(word_input, requires_grad=False), \
Variable(char_input, requires_grad=False),\
Variable(targets)
inputs = (word_input, char_input)
outputs = self.model(inputs)
# Create mask
if self.use_elmo:
mask = Variable(embeddings['mask'].transpose(0,1))
if self.use_cuda:
mask = mask.cuda()
else:
mask = create_mask(sequence_lengths, targets, cuda=self.use_cuda)
# Get CRF Loss
loss = -1*self.criterion(outputs, targets, mask=mask)
# Callbacks
test_loss += loss.item()
predictions = self.criterion.decode(outputs, mask=mask)
masked_targets = mask_targets(targets, sequence_lengths)
# Measure entity accuracy
for lab, lab_pred in zip(masked_targets, predictions):
accs += [1 if a==b else 0 for (a, b) in zip(lab, lab_pred)]
lab_chunks = set(get_chunks(lab, self.config.vocab_tags))
lab_pred_chunks = set(get_chunks(lab_pred,
self.config.vocab_tags))
correct_preds += len(lab_chunks & lab_pred_chunks)
total_preds += len(lab_pred_chunks)
total_correct += len(lab_chunks)
p = correct_preds / total_preds if correct_preds > 0 else 0
r = correct_preds / total_correct if correct_preds > 0 else 0
f1 = 2 * p * r / (p + r) if correct_preds > 0 else 0
acc = np.mean(accs)
self.logger.info("Val Loss : %.3f, Val Accuracy: %.3f%%, Val F1: %.3f%%" %(test_loss/(total_step+1), 100*acc, 100*f1))
return 100*f1
def evaluate(self,test):
batch_size = self.config.batch_size
nbatches_test, test_generator = self.batch_iter(test, batch_size,
return_lengths=True)
self.logger.info('Evaluating on test set')
self.test(nbatches_test, test_generator)
def predict_batch(self, words):
self.model.eval()
if len(words) == 1:
mult = np.ones(2).reshape(2, 1).astype(int)
if self.use_elmo:
sentences = words
character_ids = batch_to_ids(sentences)
if self.use_cuda:
character_ids = character_ids.cuda()
embeddings = self.elmo(character_ids)
word_input = embeddings['elmo_representations'][1]
word_input = Variable(word_input, requires_grad=False)
if len(words) == 1:
word_input = ((mult*word_input.transpose(0,1)).transpose(0,1).contiguous()).type(torch.FloatTensor)
word_input = T(word_input, cuda=self.use_cuda)
inputs = (word_input)
else:
#char_ids, word_ids = zip(*words)
char_ids = [[c[0] for c in s] for s in words]
word_ids = [[x[1] for x in s] for s in words]
word_ids, sequence_lengths = pad_sequences(word_ids, 1)
char_ids, word_lengths = pad_sequences(char_ids, pad_tok=0,
nlevels=2)
word_ids = np.asarray(word_ids)
char_ids = np.asarray(char_ids)
if len(words) == 1:
word_ids = mult*word_ids
char_ids = (mult*char_ids.transpose(1,0,2)).transpose(1,0,2)
word_input = T(word_ids, cuda=self.use_cuda)
char_input = T(char_ids, cuda=self.use_cuda)
word_input, char_input = Variable(word_input, requires_grad=False), \
Variable(char_input, requires_grad=False)
inputs = (word_input, char_input)
outputs = self.model(inputs)
predictions = self.criterion.decode(outputs)
predictions = [p[:i] for p, i in zip(predictions, sequence_lengths)]
return predictions
def predict(self, sentences):
"""Returns list of tags Args: words_raw: list of words (string), just one sentence (no batch) Returns: preds: list of tags (string), one for each word in the sentence """
nlp = spacy.load('en')
doc = nlp(sentences)
words_raw = [[token.text for token in sent] for sent in doc.sents]
if self.use_elmo:
words = words_raw
else:
words = [[self.config.processing_word(w) for w in s] for s in words_raw]
# print(words)
# raise NameError('testing')
# if type(words[0]) == tuple:
# words = zip(*words)
pred_ids = self.predict_batch(words)
preds = [[self.idx_to_tag[idx.item() if isinstance(idx, torch.Tensor) else idx] for idx in s] for s in pred_ids]
return preds
def create_mask(sequence_lengths, targets, cuda, batch_first=False):
""" Creates binary mask """
mask = Variable(torch.ones(targets.size()).type(torch.ByteTensor))
if cuda: mask = mask.cuda()
for i,l in enumerate(sequence_lengths):
if batch_first:
if l < targets.size(1):
mask.data[i, l:] = 0
else:
if l < targets.size(0):
mask.data[l:, i] = 0
return mask
def mask_targets(targets, sequence_lengths, batch_first=False):
""" Masks the targets """
if not batch_first:
targets = targets.transpose(0,1)
t = []
for l, p in zip(targets,sequence_lengths):
t.append(l[:p].data.tolist())
return t
6、 ... and 、 Calculation loss value (CRF)
Emission matrix :



give an example :

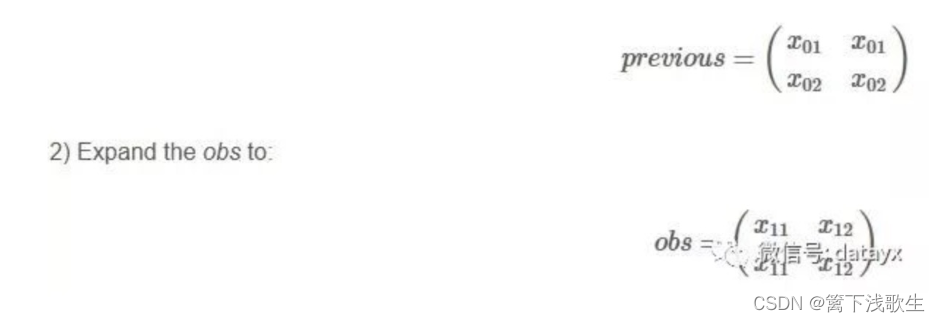

crf.py
from typing import List, Optional, Union
from torch.autograd import Variable
import torch
import torch.nn as nn
# CRF Loss function
# Function call :self.criterion = CRF(self.config.ntags)
class CRF(nn.Module):
"""Conditional random field. This module implements a conditional random field [LMP]. The forward computation of this class computes the log likelihood of the given sequence of tags and emission score tensor. This class also has ``decode`` method which finds the best tag sequence given an emission score tensor using `Viterbi algorithm`_. Arguments --------- num_tags : int Number of tags. Attributes ---------- num_tags : int Number of tags passed to ``__init__``. start_transitions : :class:`~torch.nn.Parameter` Start transition score tensor of size ``(num_tags,)``. end_transitions : :class:`~torch.nn.Parameter` End transition score tensor of size ``(num_tags,)``. transitions : :class:`~torch.nn.Parameter` Transition score tensor of size ``(num_tags, num_tags)``. References ---------- .. [LMP] Lafferty, J., McCallum, A., Pereira, F. (2001). "Conditional random fields: Probabilistic models for segmenting and labeling sequence data". *Proc. 18th International Conf. on Machine Learning*. Morgan Kaufmann. pp. 282–289. .. _Viterbi algorithm: https://en.wikipedia.org/wiki/Viterbi_algorithm """
def __init__(self, num_tags: int) -> None:
if num_tags <= 0:
raise ValueError(f'invalid number of tags: {
num_tags}')
super().__init__()
self.num_tags = num_tags
self.start_transitions = nn.Parameter(torch.Tensor(num_tags))
self.end_transitions = nn.Parameter(torch.Tensor(num_tags))
self.transitions = nn.Parameter(torch.Tensor(num_tags, num_tags))
self.reset_parameters()
def reset_parameters(self) -> None:
"""Initialize the transition parameters. The parameters will be initialized randomly from a uniform distribution between -0.1 and 0.1. """
nn.init.uniform(self.start_transitions, -0.1, 0.1)
nn.init.uniform(self.end_transitions, -0.1, 0.1)
nn.init.uniform(self.transitions, -0.1, 0.1)
def __repr__(self) -> str:
return f'{
self.__class__.__name__}(num_tags={
self.num_tags})'
def forward(self,
emissions: Variable,
tags: Variable,
mask: Optional[Variable] = None,
reduce: bool = True,
) -> Variable:
"""Compute the log likelihood of the given sequence of tags and emission score. Arguments --------- emissions : :class:`~torch.autograd.Variable` Emission score tensor of size ``(seq_length, batch_size, num_tags)``. tags : :class:`~torch.autograd.Variable` Sequence of tags as ``LongTensor`` of size ``(seq_length, batch_size)``. mask : :class:`~torch.autograd.Variable`, optional Mask tensor as ``ByteTensor`` of size ``(seq_length, batch_size)``. reduce : bool Whether to sum the log likelihood over the batch. Returns ------- :class:`~torch.autograd.Variable` The log likelihood. This will have size (1,) if ``reduce=True``, ``(batch_size,)`` otherwise. """
if emissions.dim() != 3:
raise ValueError(f'emissions must have dimension of 3, got {
emissions.dim()}')
if tags.dim() != 2:
raise ValueError(f'tags must have dimension of 2, got {
tags.dim()}')
if emissions.size()[:2] != tags.size():
raise ValueError(
'the first two dimensions of emissions and tags must match, '
f'got {
tuple(emissions.size()[:2])} and {
tuple(tags.size())}'
)
if emissions.size(2) != self.num_tags:
raise ValueError(
f'expected last dimension of emissions is {
self.num_tags}, '
f'got {
emissions.size(2)}'
)
if mask is not None:
if tags.size() != mask.size():
raise ValueError(
f'size of tags and mask must match, got {
tuple(tags.size())} '
f'and {
tuple(mask.size())}'
)
if not all(mask[0].data):
raise ValueError('mask of the first timestep must all be on')
if mask is None:
mask = Variable(self._new(tags.size()).fill_(1)).byte()
# Calculate the transfer matrix and emission matrix of the real path
numerator = self._compute_joint_llh(emissions, tags, mask)
# Calculate the of other paths log value
denominator = self._compute_log_partition_function(emissions, mask)
# obtain loss
llh = numerator - denominator
return llh if not reduce else torch.sum(llh)
def decode(self,
emissions: Union[Variable, torch.FloatTensor],
mask: Optional[Union[Variable, torch.ByteTensor]] = None) -> List[List[int]]:
"""Find the most likely tag sequence using Viterbi algorithm. Arguments --------- emissions : :class:`~torch.autograd.Variable` or :class:`~torch.FloatTensor` Emission score tensor of size ``(seq_length, batch_size, num_tags)``. mask : :class:`~torch.autograd.Variable` or :class:`torch.ByteTensor` Mask tensor of size ``(seq_length, batch_size)``. Returns ------- list List of list containing the best tag sequence for each batch. """
if emissions.dim() != 3:
raise ValueError(f'emissions must have dimension of 3, got {
emissions.dim()}')
if emissions.size(2) != self.num_tags:
raise ValueError(
f'expected last dimension of emissions is {
self.num_tags}, '
f'got {
emissions.size(2)}'
)
if mask is not None and emissions.size()[:2] != mask.size():
raise ValueError(
'the first two dimensions of emissions and mask must match, '
f'got {
tuple(emissions.size()[:2])} and {
tuple(mask.size())}'
)
if isinstance(emissions, Variable):
emissions = emissions.data
if mask is None:
mask = self._new(emissions.size()[:2]).fill_(1).byte()
elif isinstance(mask, Variable):
mask = mask.data
return self._viterbi_decode(emissions, mask)
# Function call :numerator = self._compute_joint_llh(emissions, tags, mask)
# Calculate the transfer matrix and emission matrix of the real path
def _compute_joint_llh(self,
emissions: Variable,
tags: Variable,
mask: Variable) -> Variable:
# emissions: (seq_length, batch_size, num_tags)
# tags: (seq_length, batch_size)
# mask: (seq_length, batch_size)
assert emissions.dim() == 3 and tags.dim() == 2
assert emissions.size()[:2] == tags.size()
assert emissions.size(2) == self.num_tags
assert mask.size() == tags.size()
assert all(mask[0].data)
seq_length = emissions.size(0)
mask = mask.float()
# Start transition score
llh = self.start_transitions[tags[0]] # (batch_size,)
for i in range(seq_length - 1):
cur_tag, next_tag = tags[i], tags[i+1]
# Emission score for current tag
llh += emissions[i].gather(1, cur_tag.view(-1, 1)).squeeze(1) * mask[i]
# Transition score to next tag
transition_score = self.transitions[cur_tag, next_tag]
# Only add transition score if the next tag is not masked (mask == 1)
llh += transition_score * mask[i+1]
# Find last tag index
last_tag_indices = mask.long().sum(0) - 1 # (batch_size,)
last_tags = tags.gather(0, last_tag_indices.view(1, -1)).squeeze(0)
# End transition score
llh += self.end_transitions[last_tags]
# Emission score for the last tag, if mask is valid (mask == 1)
llh += emissions[-1].gather(1, last_tags.view(-1, 1)).squeeze(1) * mask[-1]
return llh
# Calculate the of other paths log value
def _compute_log_partition_function(self,
emissions: Variable,
mask: Variable) -> Variable:
# emissions: (seq_length, batch_size, num_tags)
# mask: (seq_length, batch_size)
assert emissions.dim() == 3 and mask.dim() == 2
assert emissions.size()[:2] == mask.size()
assert emissions.size(2) == self.num_tags
assert all(mask[0].data)
seq_length = emissions.size(0)
mask = mask.float()
# Start transition score and first emission
# cache
log_prob = self.start_transitions.view(1, -1) + emissions[0]
# Here, log_prob has size (batch_size, num_tags) where for each batch,
# the j-th column stores the log probability that the current timestep has tag j
for i in range(1, seq_length):
# Broadcast log_prob over all possible next tags
# Cache matrix
broadcast_log_prob = log_prob.unsqueeze(2) # (batch_size, num_tags, 1)
# Broadcast transition score over all instances in the batch
# transition matrix
broadcast_transitions = self.transitions.unsqueeze(0) # (1, num_tags, num_tags)
# Broadcast emission score over all possible current tags
# Emission matrix
broadcast_emissions = emissions[i].unsqueeze(1) # (batch_size, 1, num_tags)
# Sum current log probability, transition, and emission scores
score = broadcast_log_prob + broadcast_transitions \
+ broadcast_emissions # (batch_size, num_tags, num_tags)
# Sum over all possible current tags, but we're in log prob space, so a sum
# becomes a log-sum-exp
score = self._log_sum_exp(score, 1) # (batch_size, num_tags)
# Set log_prob to the score if this timestep is valid (mask == 1), otherwise
# leave it alone
log_prob = score * mask[i].unsqueeze(1) + log_prob * (1.-mask[i]).unsqueeze(1)
# End transition score
log_prob += self.end_transitions.view(1, -1)
# Sum (log-sum-exp) over all possible tags
return self._log_sum_exp(log_prob, 1) # (batch_size,)
def _viterbi_decode(self, emissions: torch.FloatTensor, mask: torch.ByteTensor) \
-> List[List[int]]:
# Get input sizes
seq_length = emissions.size(0)
batch_size = emissions.size(1)
sequence_lengths = mask.long().sum(dim=0)
# emissions: (seq_length, batch_size, num_tags)
assert emissions.size(2) == self.num_tags
# list to store the decoded paths
best_tags_list = []
# Start transition
viterbi_score = []
viterbi_score.append(self.start_transitions.data + emissions[0])
viterbi_path = []
# Here, viterbi_score is a list of tensors of shapes of (num_tags,) where value at
# index i stores the score of the best tag sequence so far that ends with tag i
# viterbi_path saves where the best tags candidate transitioned from; this is used
# when we trace back the best tag sequence
# Viterbi algorithm recursive case: we compute the score of the best tag sequence
# for every possible next tag
for i in range(1, seq_length):
# Broadcast viterbi score for every possible next tag
broadcast_score = viterbi_score[i - 1].view(batch_size, -1, 1)
# Broadcast emission score for every possible current tag
broadcast_emission = emissions[i].view(batch_size, 1, -1)
# Compute the score matrix of shape (batch_size, num_tags, num_tags) where
# for each sample, each entry at row i and column j stores the score of
# transitioning from tag i to tag j and emitting
score = broadcast_score + self.transitions.data + broadcast_emission
# Find the maximum score over all possible current tag
best_score, best_path = score.max(1) # (batch_size,num_tags,)
# Save the score and the path
viterbi_score.append(best_score)
viterbi_path.append(best_path)
# Now, compute the best path for each sample
for idx in range(batch_size):
# Find the tag which maximizes the score at the last timestep; this is our best tag
# for the last timestep
seq_end = sequence_lengths[idx]-1
_, best_last_tag = (viterbi_score[seq_end][idx] + self.end_transitions.data).max(0)
best_tags = [best_last_tag.item()] #[best_last_tag[0]] #[best_last_tag.item()]
# We trace back where the best last tag comes from, append that to our best tag
# sequence, and trace it back again, and so on
for path in reversed(viterbi_path[:sequence_lengths[idx] - 1]):
best_last_tag = path[idx][best_tags[-1]]
best_tags.append(best_last_tag)
# Reverse the order because we start from the last timestep
best_tags.reverse()
best_tags_list.append(best_tags)
return best_tags_list
@staticmethod
def _log_sum_exp(tensor: Variable, dim: int) -> Variable:
# Find the max value along `dim`
offset, _ = tensor.max(dim)
# Make offset broadcastable
broadcast_offset = offset.unsqueeze(dim)
# Perform log-sum-exp safely
safe_log_sum_exp = torch.log(torch.sum(torch.exp(tensor - broadcast_offset), dim))
# Add offset back
return offset + safe_log_sum_exp
def _new(self, *args, **kwargs) -> torch.FloatTensor:
param = next(self.parameters())
return param.data.new(*args, **kwargs)
7、 ... and 、 Training (train.py)
from model.data_utils import CoNLLDataset
from model.config import Config
from model.ner_model import NERModel
from model.ner_learner import NERLearner
def main():
# create instance of config
config = Config()
if config.use_elmo: config.processing_word = None
#build model
# NERModel Model
model = NERModel(config)
# create datasets
# Training data preprocessing
dev = CoNLLDataset(config.filename_dev, config.processing_word,
config.processing_tag, config.max_iter, config.use_crf)
train = CoNLLDataset(config.filename_train, config.processing_word,
config.processing_tag, config.max_iter, config.use_crf)
# Initialize the model training process
learn = NERLearner(config, model)
# 1. according to batch_size Collate validation data sets
# 2. The incoming to elmo gain input
# 3. The incoming to NERModel In the model
# 4. use CRF seek loss value
learn.fit(train, dev)
if __name__ == "__main__":
main()
8、 ... and 、 test (test.py)
""" Command Line Usage Args: eval: Evaluate F1 Score and Accuracy on test set pred: Predict sentence. (optional): Sentence to predict on. If none given, predicts on "Peter Johnson lives in Los Angeles" Example: > python test.py eval pred "Obama is from Hawaii" """
from model.data_utils import CoNLLDataset
from model.config import Config
from model.ner_model import NERModel
from model.ner_learner import NERLearner
import sys
def main():
# create instance of config
config = Config()
if config.use_elmo: config.processing_word = None
#build model
model = NERModel(config)
learn = NERLearner(config, model)
learn.load()
if len(sys.argv) == 1:
print("No arguments given. Running full test")
sys.argv.append("eval")
sys.argv.append("pred")
if sys.argv[1] == "eval":
# create datasets
test = CoNLLDataset(config.filename_test, config.processing_word,
config.processing_tag, config.max_iter)
learn.evaluate(test)
if sys.argv[1] == "pred" or sys.argv[2] == "pred":
try:
sent = (sys.argv[2] if sys.argv[1] == "pred" else sys.argv[3])
except IndexError:
sent = ["Peter", "Johnson", "lives", "in", "Los", "Angeles"]
print("Predicting sentence: ", sent)
pred = learn.predict(sent)
print(pred)
if __name__ == "__main__":
main()
experimental result
obtain loss value

版权声明
本文为[Shallow singing under the hedge]所创,转载请带上原文链接,感谢
https://yzsam.com/2022/04/202204231349535417.html
边栏推荐
- The query did not generate a result set exception resolution when the dolphin scheduler schedules the SQL task to create a table
- Oracle clear SQL cache
- Dolphin scheduler source package Src tar. GZ decompression problem
- 3300万IOPS、39微秒延迟、碳足迹认证,谁在认真搞事情?
- Express中间件③(自定义中间件)
- Using Baidu Intelligent Cloud face detection interface to achieve photo quality detection
- Oracle renames objects
- Storage scheme of video viewing records of users in station B
- PG library checks the name
- UNIX final exam summary -- for direct Department
猜你喜欢
The art of automation
专题测试05·二重积分【李艳芳全程班】

OSS cloud storage management practice (polite experience)

Android interview theme collection

Oracle defines self incrementing primary keys through triggers and sequences, and sets a scheduled task to insert a piece of data into the target table every second

Express middleware ③ (custom Middleware)

Dolphin scheduler configuring dataX pit records

SQL learning | set operation
What is the difference between blue-green publishing, rolling publishing and gray publishing?

Small case of web login (including verification code login)
随机推荐
[code analysis (3)] communication efficient learning of deep networks from decentralized data
RAC environment error reporting ora-00239: timeout waiting for control file enqueue troubleshooting
Reading notes: meta matrix factorization for federated rating predictions
AtomicIntegerArray源码分析与感悟
L2-024 部落 (25 分)
[code analysis (6)] communication efficient learning of deep networks from decentralized data
Troubleshooting of expdp export error when Oracle table has logical bad blocks
Technologie zéro copie
Port occupied 1
Window function row commonly used for fusion and de duplication_ number
2021年秋招,薪资排行NO
[VMware] address of VMware Tools
Dolphin scheduler configuring dataX pit records
JMeter pressure test tool
Express中间件③(自定义中间件)
19c environment ora-01035 login error handling
Detailed explanation and usage of with function in SQL
MySQL [read / write lock + table lock + row lock + mvcc]
【vmware】vmware tools 地址
Storage scheme of video viewing records of users in station B